Coregistration with SPM and RHINO
This tutorial covers the registration of MRI and M/EEG datasets into a common space to allow for analysis in sourcespace.
This practical uses data from the coreg_example subfolders in the OSL example_data directory. Please make sure this is present before starting.
Contents
Coordinate Systems
There are several co-ordinate systems which must be aligned before we can project our sensor MEG data into source space. These are:
- Scanner Coordinates - These are the locations of MEG sensors within the dewar.
- Polhemus Coordinates - These include the relative locations of the Fiducial Locations (LPA, RPA and Nasion), Head Position Indicator Coils and headshape points. These are acquired prior to the MEG scan.
- MRI Coordinates - This is the information from the individuals structural T1 MRI scan.
See https://ohba-analysis.github.io/osl-docs/pages/docs/preprocessing.html#coregistration-using-rhino for more information.
Coregistration summary
The coregistration follows these steps
- Extraction of the scalp shape and fiducial points from the MRI scan.
- Alignment of the MRI and Polhemus data, first by fiducials and refined by the scalp and headshape points.
- Positioning the Polhemus and MRI coordinates within the sensors using the Head Position Indicator Coils.
Once these transforms have been identified we can move between MEG sensors and specific locations within the MRI scan.
Setup the analysis
To start OSL only thing you need to do is to go into your OSL directory (i.e. type cd /somedirectory/osl-core ) and then run the following.
osl_startup;
This only needs to be run once, each time you re-start MATLAB
We now setup the location of the data to be coregistered
datadir = fullfile(osldir,'example_data','coreg_example'); spm_files_continuous=[datadir '/Aface_meg1.mat'];
Coregistration using SPM
Coregistration is performed in OSL using osl_headmodel.
We will first run a standard coregistration using SPM. This takes an option structure defining several key parameters:
- S.D - spm object filename
- S.mri - structural mri scan filename
- S.useheadshape - set to 0 or 1 to indicate whether to use the Polhemus headshape points to refine the alignment between the MRI and Polhemus data
The following example runs standard SPM coregistration on an example dataset. While running, SPM will open a window showing the alignment of the headshape points to the scalp.
S = []; S.D = spm_files_continuous; S.mri = fullfile(osldir,'example_data','faces_singlesubject','structurals','struct.nii'); S.useheadshape = 1; S.use_rhino = 0; S.forward_meg = 'Single Shell'; S.fid.label.nasion = 'Nasion'; S.fid.label.lpa = 'LPA'; S.fid.label.rpa = 'RPA'; D=osl_headmodel(S);
View SPM registration
This SPM tool allows us to view the results of the coregistration (click and drag to rotate the image - you may need to click on the "Rotate 3D" toolbar button)
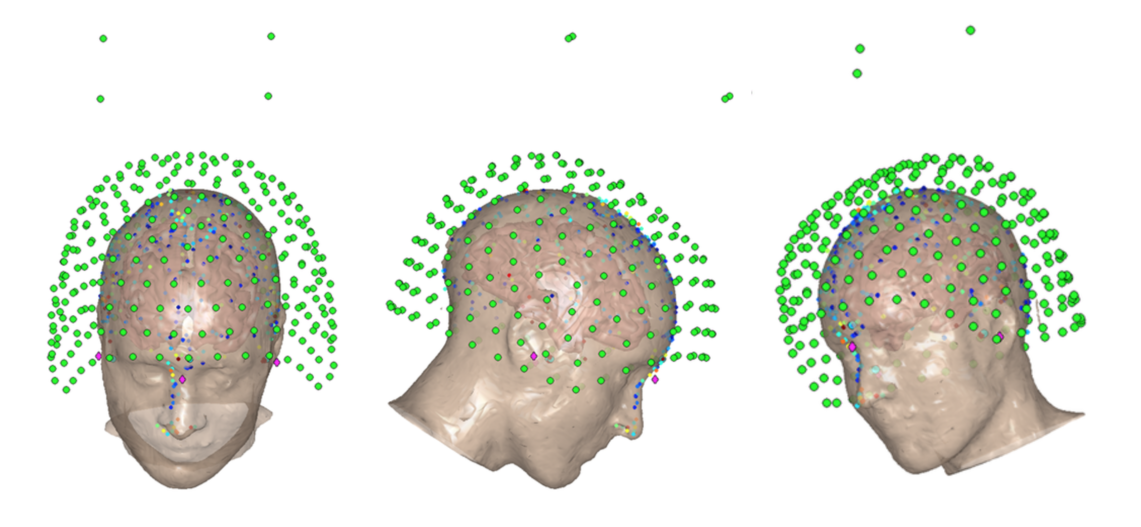
The coregistration shows several types of information:
- Green circles - MEG sensors (from an Elekta System)
- Pink Diamonds - Fiducial locations (LPA, RPA and Nasion)
- Light Red Surfact - Scalp extraction
- Red Surface - Inner skull extraction
- Blue Surface - Brain surface
- Tiny Red Dots - Headshape points
figure('Position',[100 100 1024 1024])
spm_eeg_inv_checkdatareg_3Donly(spm_files_continuous);
zoom(0.5)
Coregistration with RHINO
To ensure a good coregistration we must make sure that the alignment between the MRI and Polhemus coordinates is as accurate as possible. There are several things we can do to improve on the typical pipeline:
- Having a large number of Polhemus headshape points (>100) to avoid local minima in the alignment.
- Including the nose in the MRI surface extraction and Polhemus headshape points. As the head is approximately spherical, it is easy to get a rotational error if we just use the scalp. By including parts of the face and nose, we can improve the alignment.
RHINO is an OSL tool implementing these additional improvements. You need to make sure that you have a clear nose on your MRI scan and a large number of headshape points covering the scalp and ridgid parts of the face.
datadir = fullfile(osldir,'example_data','coreg_example'); spm_file=[datadir '/pdsubject1']; S = []; S.D = spm_file; S.mri = fullfile(osldir,'example_data','coreg_example','subject1_struct_canon.nii'); S.useheadshape = 1; S.use_rhino = 1; % We now set the RHINO option to 1 S.forward_meg = 'Single Shell'; S.fid.label.nasion = 'Nasion'; S.fid.label.lpa = 'LPA'; S.fid.label.rpa = 'RPA'; D=osl_headmodel(S);
Visualising structural preprocessing
The first time RHINO coregistration is run the structural data will be preprocessed using FSL tools. These perform a linear registration, brain extraction and scalp extraction from the T1 scan.
More information about RHINO can be found here:
https://ohba-analysis.github.io/osl-docs/pages/docs/preprocessing.html#coregistration-using-rhino
We can visualise the extracted scalp surface and original MRI scan in fsleyes with the following call.
This will load the structural scan overlayed on the scalp extraction. You can turn-off the strutural scan by double clicking on the picture of the eye to the left of the label 'subject1_struct_canon.nii' in the middle-bottom of the screen. Underneath, the scalp is the boundary between the white (outside) and black (inside) regions.
Try turning the structural scan on and off whilst exploring the image. There is a good correspondance between the scalp on the MRI and the extracted scalp image. Any deformations in the extracted scalp could be misleading when we align to our headshape points. In this case, the structrual preprcessing has worked well.
mri = fullfile(osldir,'example_data','coreg_example','rhino_subject1_struct_canon.nii'); scalp = fullfile(osldir,'example_data','coreg_example','rhino_subject1_struct_canon_scalp.nii.gz'); fsleyes({mri, scalp},[],'greyscale','none')
Visualising the RHINO coregistration
We can visualise the full coregistration by calling rhino_display(D).
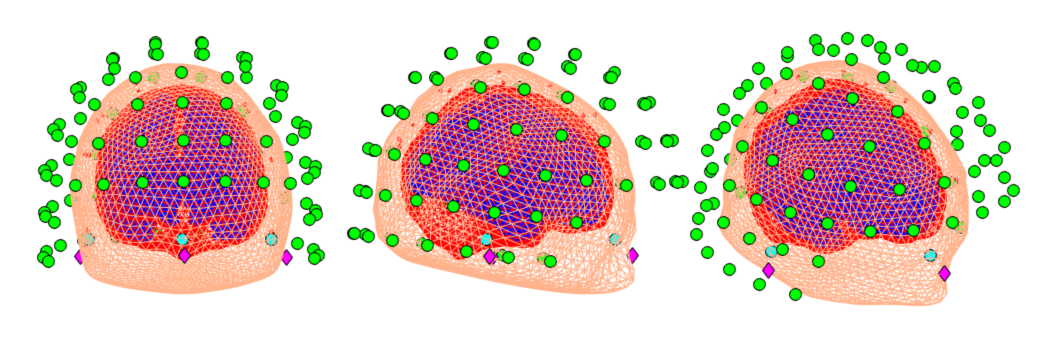
This contains similar information to the SPM coreg.
- Green Dots - MEG Sensors (from a CTF system including 4 Reference Coils)
- Magenta Diamonds - MRI Fiducial Locations
- Cyan Circles - MEG Fiducial Locations
- Beige Surface - Whole Head Scalp Extraction
- Pink Surface - Brain Extraction
- Small Blue-Red Dots - Headshape points colour coded by fit to the scalp. Blue indicates a close fit and Red a bad fit. A large number of Red headshape points indicates that the general fit might be bad.
You can click and drag the image to explore the registration.
Note the close correspondance between the headshape points and scalp extraction. The inclusion of the whole head and nose gives us greater confidence in the quality of the coregistration. Compare the RHINO surfaces with those from the SPM output.
This coregistration has worked very well. If you notice any large disparities in future analyses, these should be manually corrected at this stage:
- Bad scalp extractions can be improved by tuning the FSL BET.
- Misleading or erroneous headshape points can be removed.
- Badly estimated Fiducial locations can be reestimated.
D = spm_eeg_load(S.D); rhino_display(D);