osl-ephys
OHBA Software Library (OSL) Electrophysiology Toolbox
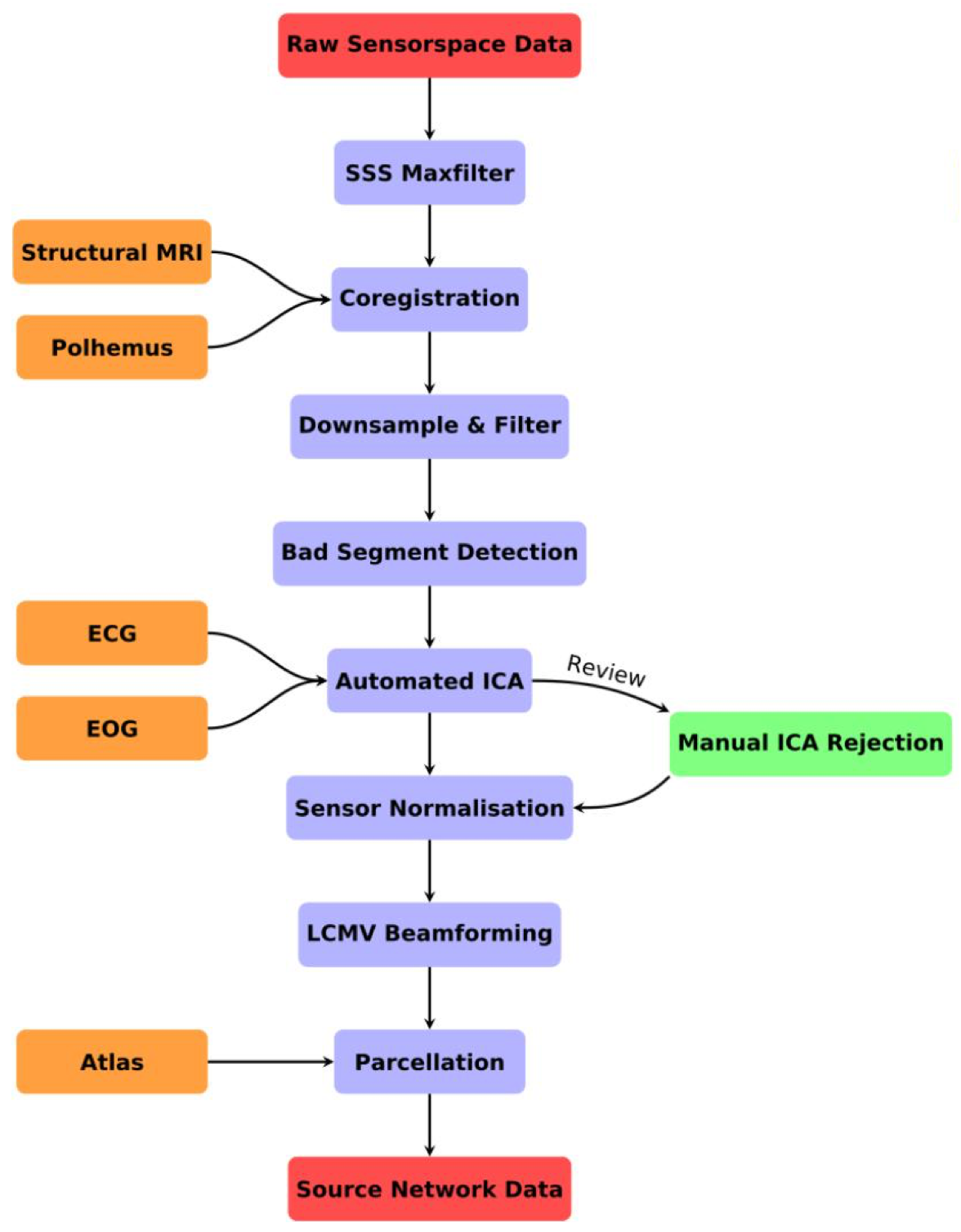
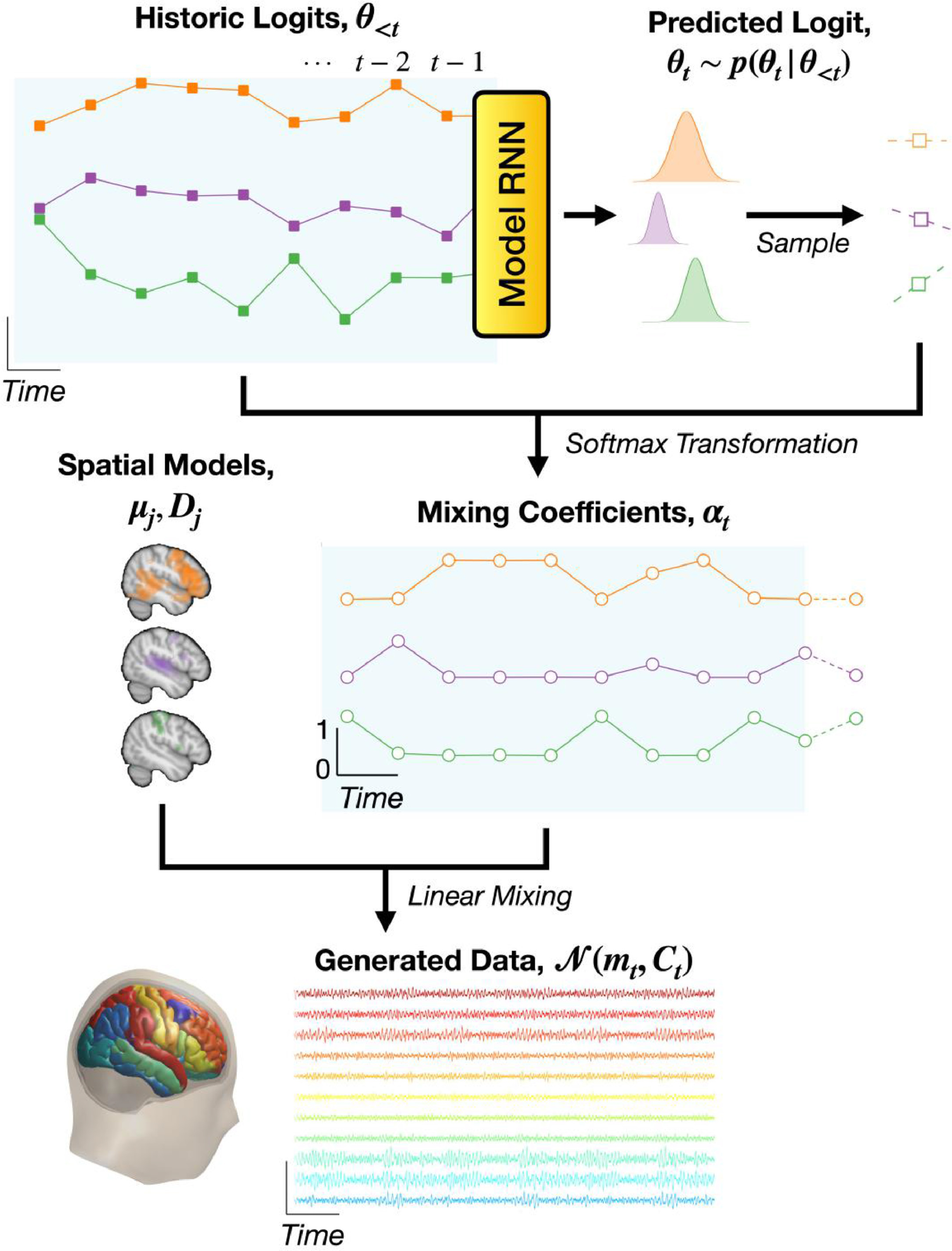
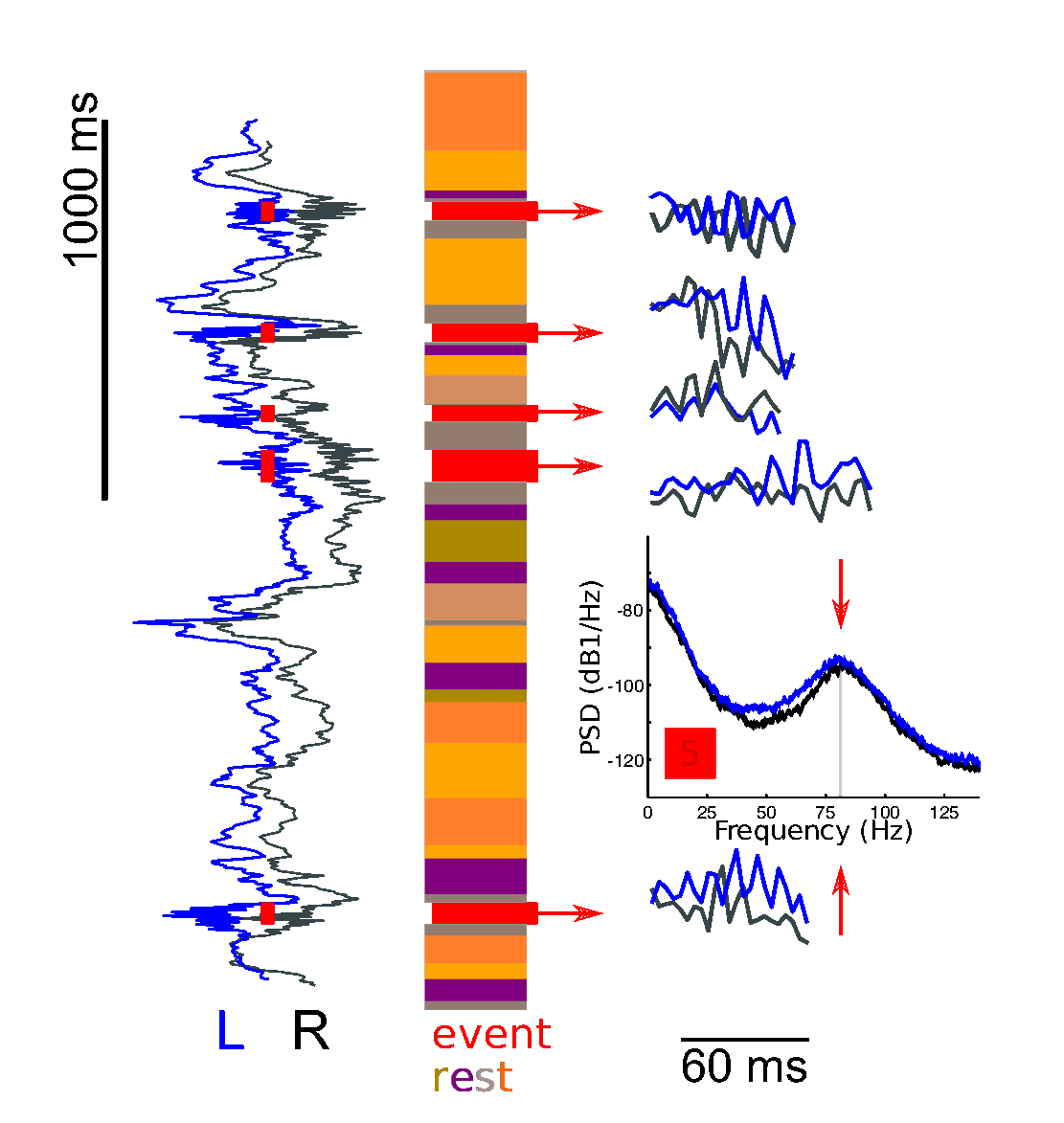
OSL-ephys is a python package built on MNE-python for the analysis of MEG and EEG data. OSL-ephys includes configurations and batching for easy parallel pipeline processing over multiple sessions of data. It also includes a purely volumetric source reconstruction pipeline using RHINO, which uses FSL to handle surfaces (as opposed to Freesurfer).